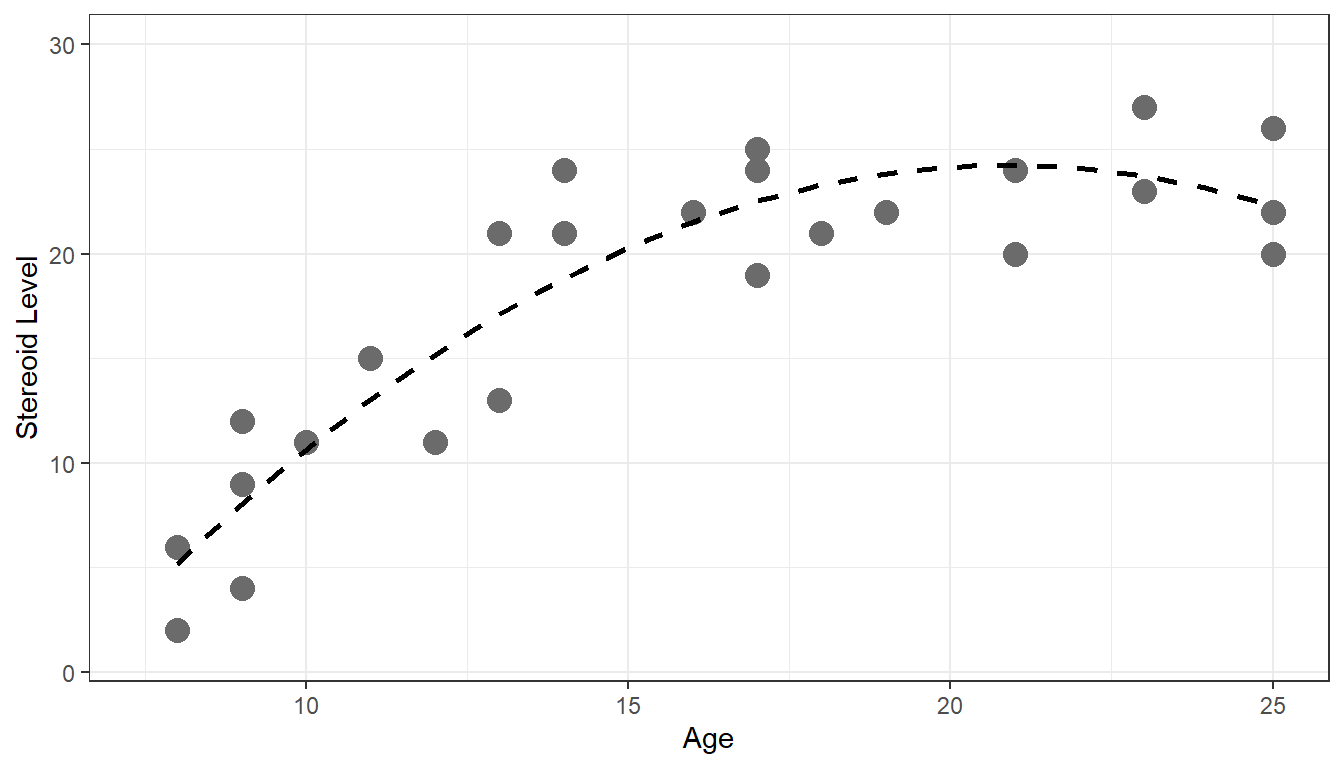1 Linear Regression with One Predictor Variable
1.1 Relations between Variables
1.1.1 Example 1
require(here) fig1.2 <- read.csv(here('/data/fig1.2.csv/'),header=FALSE) require(ggplot2) ggplot(fig1.2,aes(x=V1,y=V2))+ geom_point(size=4,col='gray42')+ geom_smooth(method=lm,se=FALSE,lty=2,col='black')+ xlim(55,100)+ ylim(55,100)+ xlab('Midyear Evaluation')+ ylab('Year-End Evaluation')+ theme_bw()
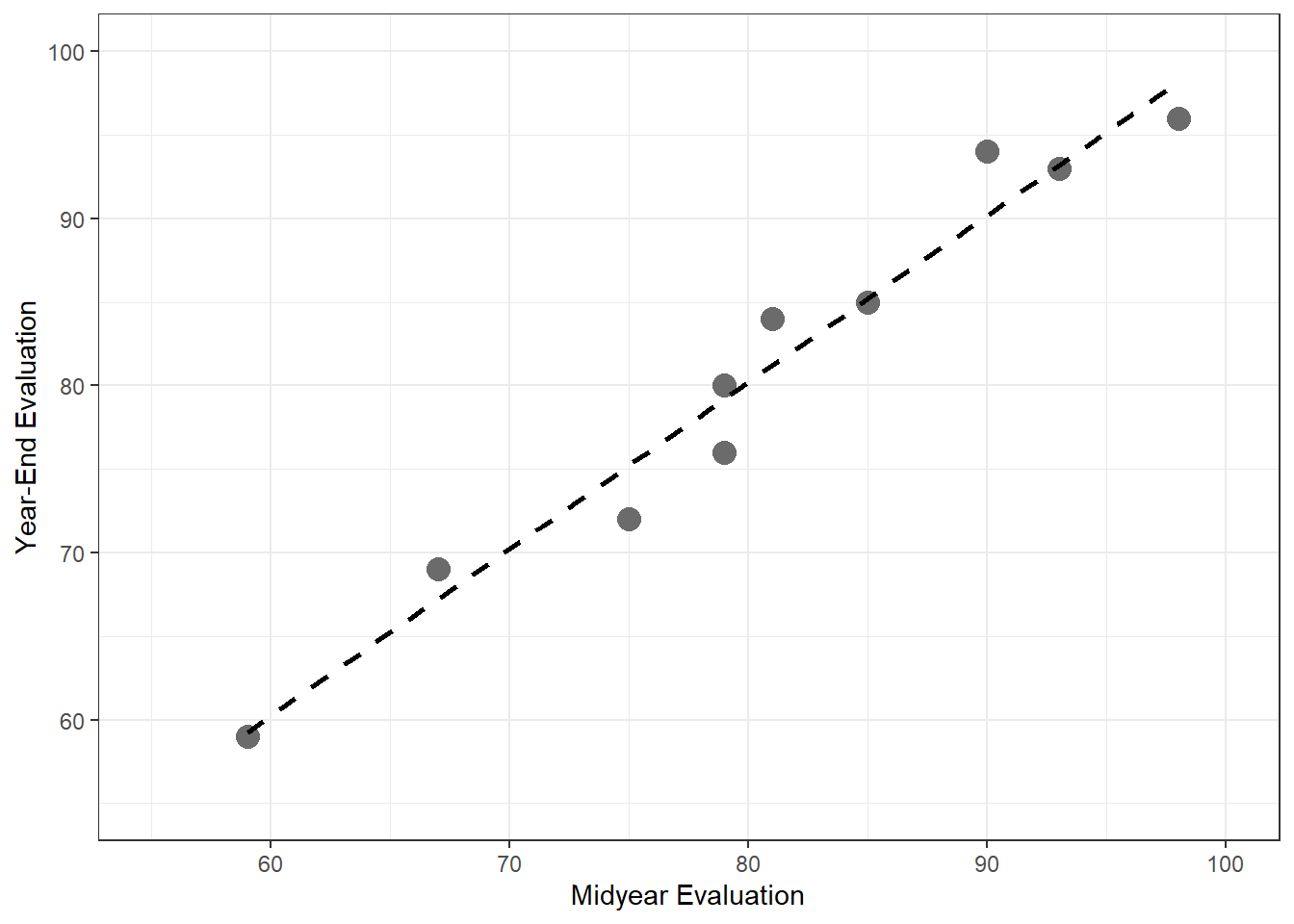
1.3 Simple Linear Regression Model with Distributions of Error Terms Unspecified
1.3.1 Example (page 10)
b0 = 9.5 b1 = 2.1 x = 0:70 mean = b0 + b1*x err.sd = 5 plot(x,mean,type="l", ylim=c(40,120), xlim=c(10,50), cex=1, pch=19, xlab="Number of Bids Prepared", ylab="Hours", xaxt='n') axis(side=1,at=c(0,25,45)) abline(b0,b1) dens = dnorm(seq(-3,3,.01),0,1) for(i in c(26,46)){ x. = x[i] - 5*dens y. = mean[i]+seq(-3,3,.01)*err.sd points(x.,y.,type="l",lty=2) abline(v=x[i],lty=2,col="gray") }
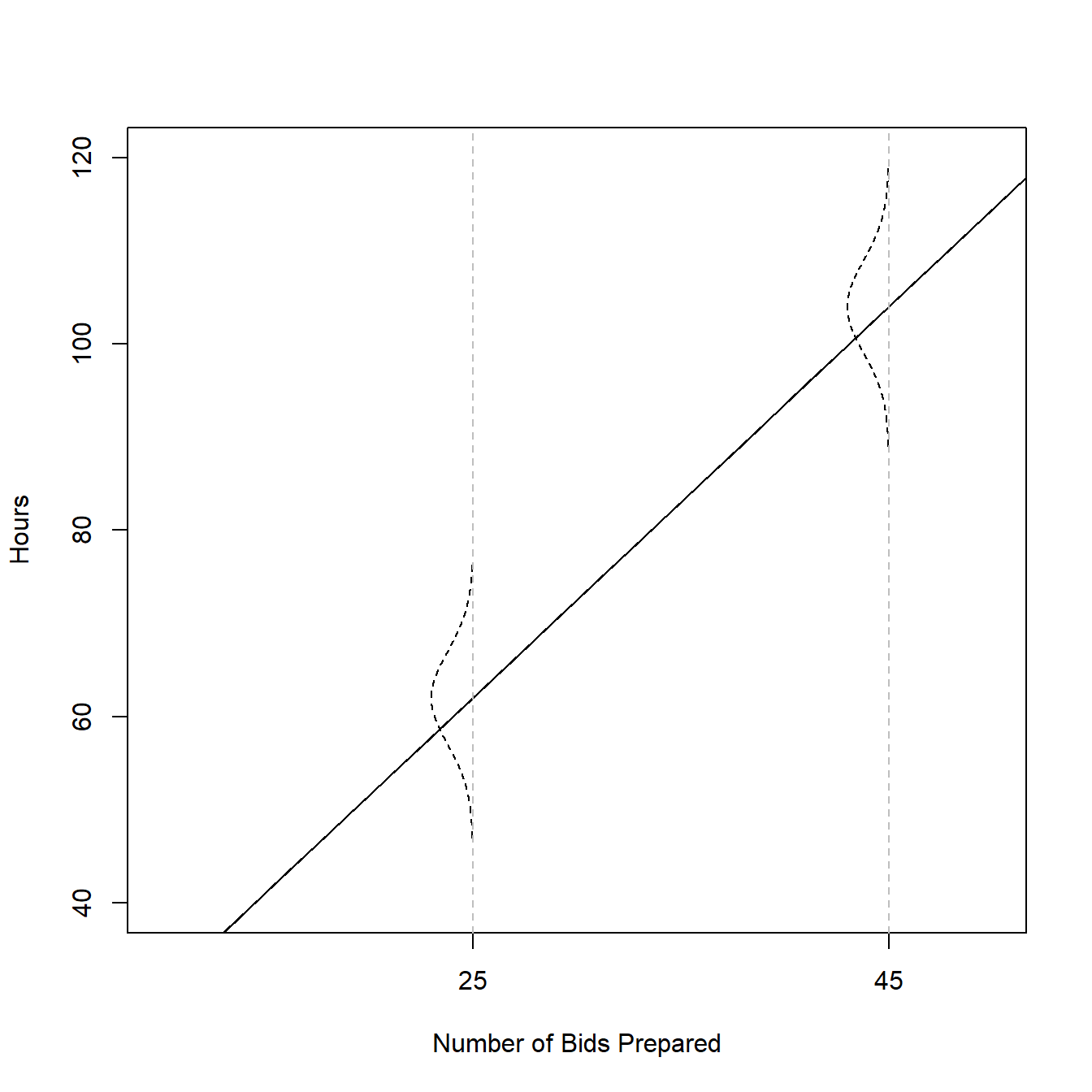
1.6 Estimation of Regression Function
1.6.1 Example (page 15)
Age <- c(20,55,30) Attempts <- c(5, 12, 10) fig1.9 <- data.frame(Age=Age,Attempts=Attempts) ggplot(data= fig1.9, aes(x=Age,y=Attempts))+ geom_point(size=4,color='gray42')+ xlim(c(10,60))+ ylim(c(0,15))+ theme_bw()+ geom_hline(yintercept = 9)+ geom_segment(x = fig1.9[1,1], y = fig1.9[1,2], xend = fig1.9[1,1], yend = 9)+ geom_segment(x = fig1.9[2,1], y = fig1.9[2,2], xend = fig1.9[2,1], yend = 9)+ geom_segment(x = fig1.9[3,1], y = fig1.9[3,2], xend = fig1.9[3,1], yend = 9)+ xlab('Age')+ ylab('Attempts')
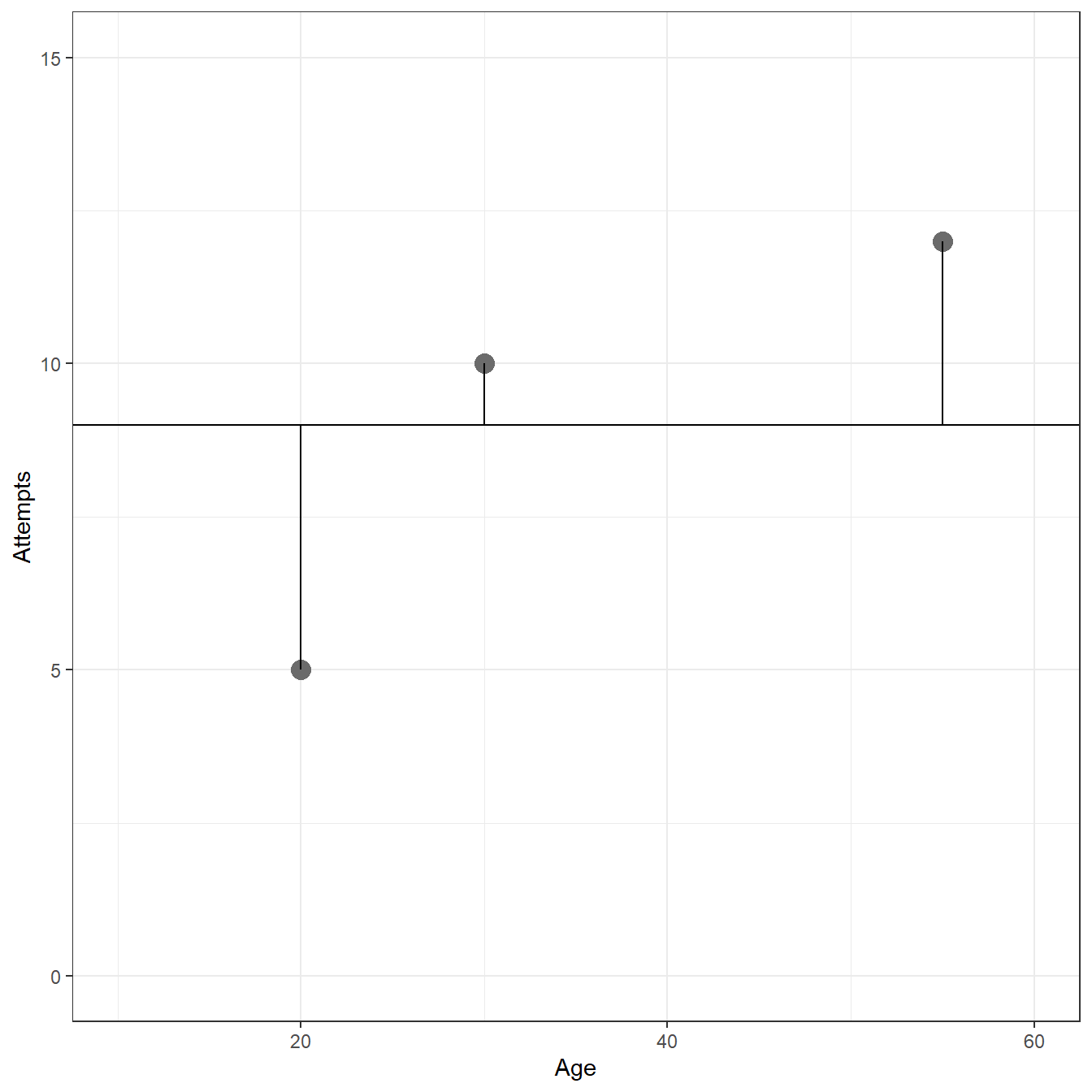
# Sum of Squared Deviations sum((Attempts - 9)^2)
[1] 26mod <- lm(Attempts ~ 1 + Age,d=fig1.9) ggplot(data= fig1.9, aes(x=Age,y=Attempts))+ geom_point(size=4,color='gray42')+ geom_abline(intercept = coef(mod)[1],slope=coef(mod)[2])+ xlim(c(10,60))+ ylim(c(0,15))+ theme_bw()+ geom_segment(x = fig1.9[1,1], y = fig1.9[1,2], xend = fig1.9[1,1], yend = predict(mod, newdata = data.frame(Age=c(fig1.9[1,1]))))+ geom_segment(x = fig1.9[2,1], y = fig1.9[2,2], xend = fig1.9[2,1], yend = predict(mod, newdata = data.frame(Age=c(fig1.9[2,1]))))+ geom_segment(x = fig1.9[3,1], y = fig1.9[3,2], xend = fig1.9[3,1], yend = predict(mod, newdata = data.frame(Age=c(fig1.9[3,1]))))+ xlab('Age')+ ylab('Attempts')
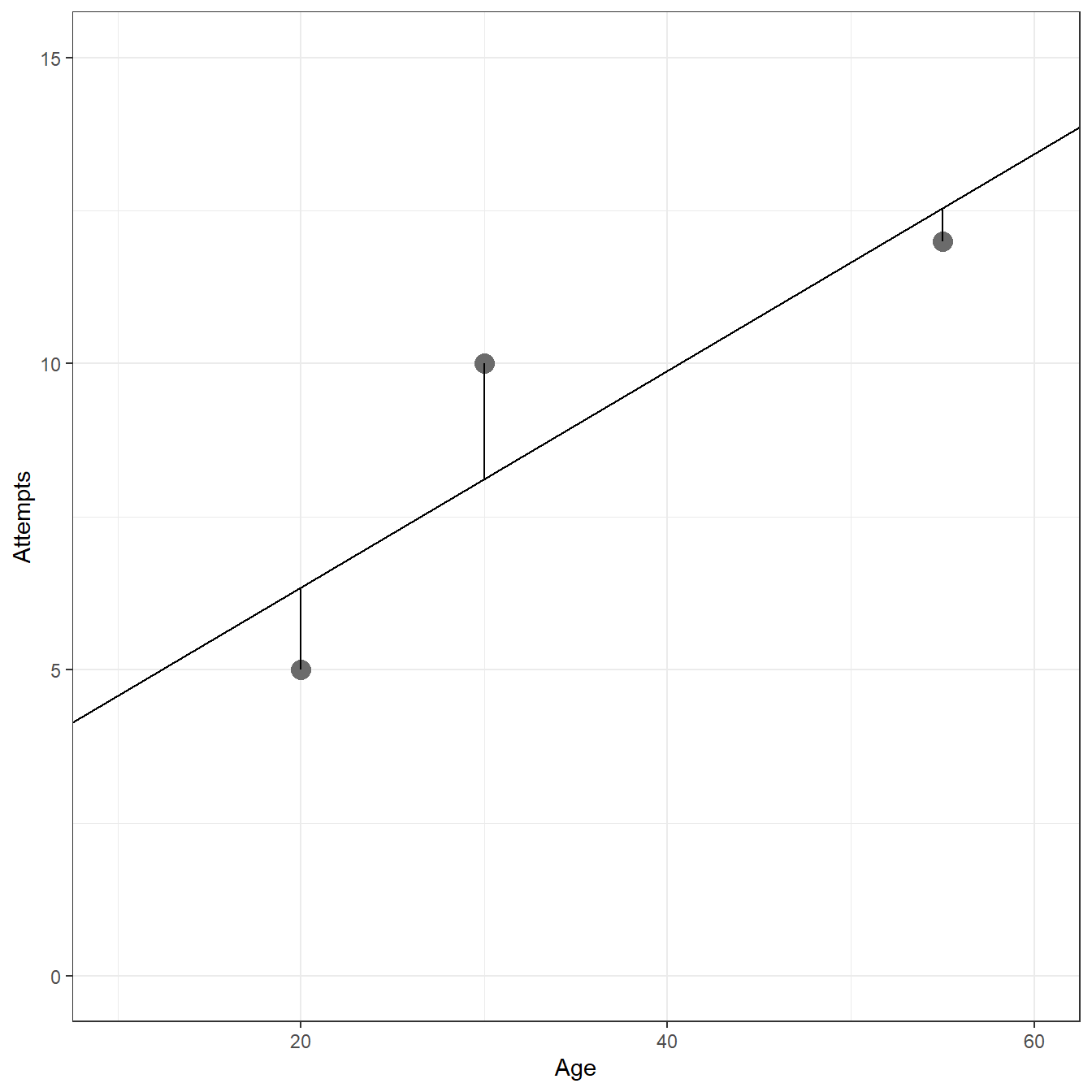
# Model predictions predict(mod)
1 2 3
6.346 12.538 8.115 # Model Residuals resid(mod)
1 2 3
-1.3462 -0.5385 1.8846 [1] 5.654[1] 5.6541.6.2 Example (page 19)
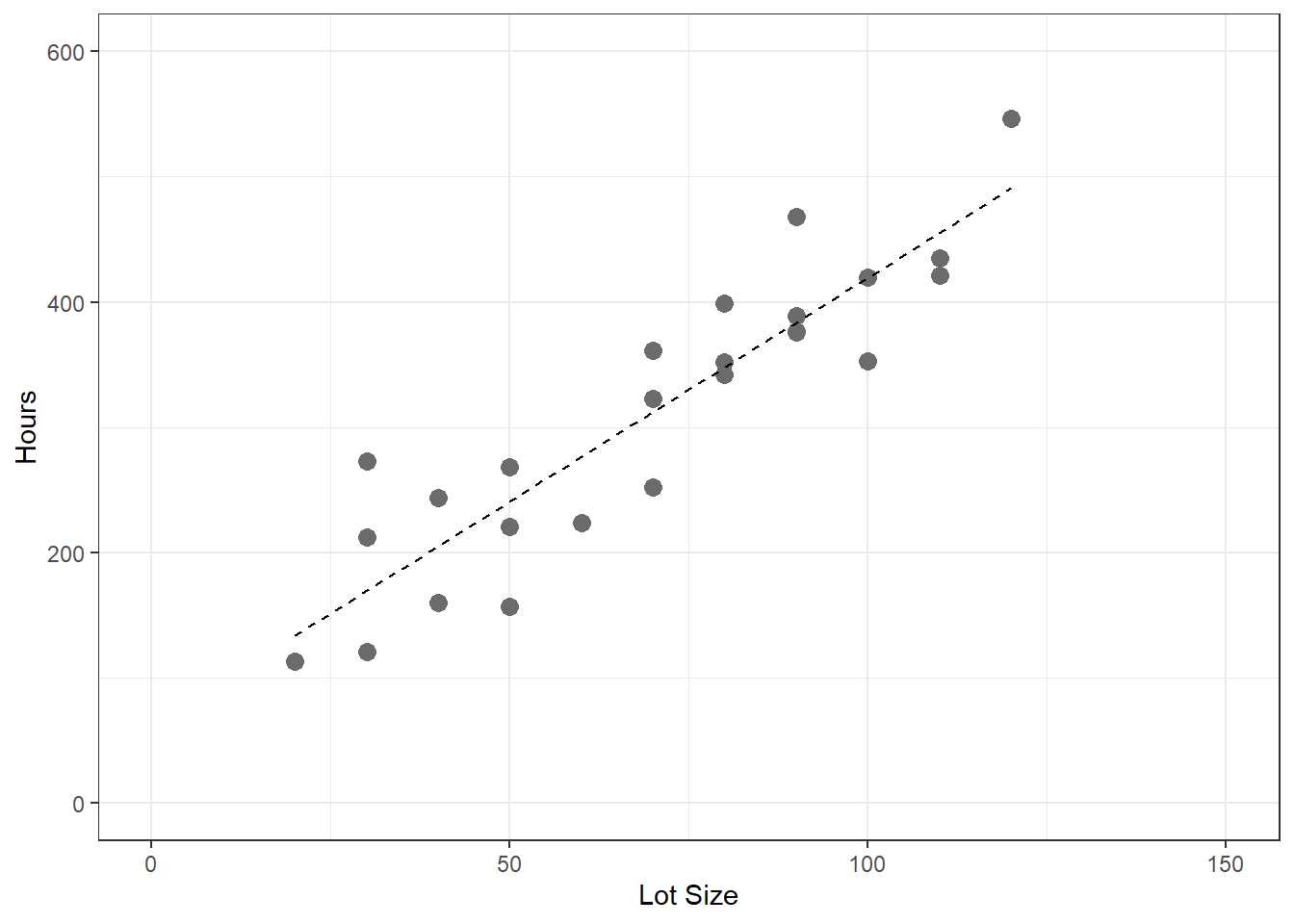
table1.1 <- read.table(here('/data/CH01TA01.txt/'),header=FALSE) colnames(table1.1) <- c('lot.size','work.hours') table1.1
lot.size work.hours
1 80 399
2 30 121
3 50 221
4 90 376
5 70 361
6 60 224
7 120 546
8 80 352
9 100 353
10 50 157
11 40 160
12 70 252
13 90 389
14 20 113
15 110 435
16 100 420
17 30 212
18 50 268
19 90 377
20 110 421
21 30 273
22 90 468
23 40 244
24 80 342
25 70 323ggplot(table1.1,aes(x=lot.size,y=work.hours))+ geom_point(size=3,col='gray42')+ geom_smooth(method=lm,se=FALSE,lty=2,lwd=0.5,col='black')+ xlim(0,150)+ ylim(0,600)+ xlab('Lot Size')+ ylab('Hours')+ theme_bw()
Anova Table (Type III tests)
Response: work.hours
Sum Sq Df F value Pr(>F)
(Intercept) 13530 1 5.68 0.026 *
lot.size 252378 1 105.88 0.00000000044 ***
Residuals 54825 23
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1summary(mod)
Call:
lm(formula = work.hours ~ 1 + lot.size, data = table1.1)
Residuals:
Min 1Q Median 3Q Max
-83.88 -34.09 -5.98 38.83 103.53
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 62.366 26.177 2.38 0.026 *
lot.size 3.570 0.347 10.29 0.00000000044 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 48.8 on 23 degrees of freedom
Multiple R-squared: 0.822, Adjusted R-squared: 0.814
F-statistic: 106 on 1 and 23 DF, p-value: 0.000000000445coef(mod)
(Intercept) lot.size
62.37 3.57 table1.1$x_xbar <- table1.1$lot.size - mean(table1.1$lot.size) table1.1$y_ybar <- table1.1$work.hours - mean(table1.1$work.hours) table1.1$x_xbar_y_ybar <- table1.1$x_xbar*table1.1$y_ybar table1.1$x_xbar.sq <- table1.1$x_xbar^2 table1.1$y_ybar.sq <- table1.1$y_ybar^2 table1.1
lot.size work.hours x_xbar y_ybar x_xbar_y_ybar x_xbar.sq y_ybar.sq
1 80 399 10 86.72 867.2 100 7520.4
2 30 121 -40 -191.28 7651.2 1600 36588.0
3 50 221 -20 -91.28 1825.6 400 8332.0
4 90 376 20 63.72 1274.4 400 4060.2
5 70 361 0 48.72 0.0 0 2373.6
6 60 224 -10 -88.28 882.8 100 7793.4
7 120 546 50 233.72 11686.0 2500 54625.0
8 80 352 10 39.72 397.2 100 1577.7
9 100 353 30 40.72 1221.6 900 1658.1
10 50 157 -20 -155.28 3105.6 400 24111.9
11 40 160 -30 -152.28 4568.4 900 23189.2
12 70 252 0 -60.28 0.0 0 3633.7
13 90 389 20 76.72 1534.4 400 5886.0
14 20 113 -50 -199.28 9964.0 2500 39712.5
15 110 435 40 122.72 4908.8 1600 15060.2
16 100 420 30 107.72 3231.6 900 11603.6
17 30 212 -40 -100.28 4011.2 1600 10056.1
18 50 268 -20 -44.28 885.6 400 1960.7
19 90 377 20 64.72 1294.4 400 4188.7
20 110 421 40 108.72 4348.8 1600 11820.0
21 30 273 -40 -39.28 1571.2 1600 1542.9
22 90 468 20 155.72 3114.4 400 24248.7
23 40 244 -30 -68.28 2048.4 900 4662.2
24 80 342 10 29.72 297.2 100 883.3
25 70 323 0 10.72 0.0 0 114.9[1] 3.57[1] 62.371.6.4 Table 1.2 (page 22)
table1.1 <- read.table(here('/data/CH01TA01.txt/'),header=FALSE) colnames(table1.1) <- c('lot.size','work.hours') mod <- lm(work.hours ~ 1 + lot.size,d=table1.1) table1.1$predicted <- predict(mod) table1.1$residuals <- resid(mod) table1.1$residuals.squared <- resid(mod)^2 table1.1
lot.size work.hours predicted residuals residuals.squared
1 80 399 348.0 51.0180 2602.8343
2 30 121 169.5 -48.4719 2349.5270
3 50 221 240.9 -19.8760 395.0538
4 90 376 383.7 -7.6840 59.0445
5 70 361 312.3 48.7200 2373.6384
6 60 224 276.6 -52.5780 2764.4440
7 120 546 490.8 55.2099 3048.1329
8 80 352 348.0 4.0180 16.1442
9 100 353 419.4 -66.3861 4407.1090
10 50 157 240.9 -83.8760 7035.1766
11 40 160 205.2 -45.1739 2040.6848
12 70 252 312.3 -60.2800 3633.6784
13 90 389 383.7 5.3160 28.2594
14 20 113 133.8 -20.7699 431.3887
15 110 435 455.1 -20.0881 403.5310
16 100 420 419.4 0.6139 0.3769
17 30 212 169.5 42.5281 1808.6377
18 50 268 240.9 27.1240 735.7136
19 90 377 383.7 -6.6840 44.6764
20 110 421 455.1 -34.0881 1161.9973
21 30 273 169.5 103.5281 10718.0635
22 90 468 383.7 84.3160 7109.1810
23 40 244 205.2 38.8261 1507.4630
24 80 342 348.0 -5.9820 35.7846
25 70 323 312.3 10.7200 114.91841.6.5 Alternative Model with Mean Centering (page 22)
table1.1 <- read.table(here('/data/CH01TA01.txt/'),header=FALSE) colnames(table1.1) <- c('lot.size','work.hours') table1.1$lot.size_centered <- table1.1$lot.size - mean(table1.1$lot.size) mod <- lm(work.hours ~ 1 + lot.size_centered,d=table1.1) Anova(mod,type=3)
Anova Table (Type III tests)
Response: work.hours
Sum Sq Df F value Pr(>F)
(Intercept) 2437970 1 1023 < 0.0000000000000002 ***
lot.size_centered 252378 1 106 0.00000000044 ***
Residuals 54825 23
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1summary(mod)
Call:
lm(formula = work.hours ~ 1 + lot.size_centered, data = table1.1)
Residuals:
Min 1Q Median 3Q Max
-83.88 -34.09 -5.98 38.83 103.53
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 312.280 9.765 32.0 < 0.0000000000000002 ***
lot.size_centered 3.570 0.347 10.3 0.00000000044 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 48.8 on 23 degrees of freedom
Multiple R-squared: 0.822, Adjusted R-squared: 0.814
F-statistic: 106 on 1 and 23 DF, p-value: 0.000000000445coef(mod)
(Intercept) lot.size_centered
312.28 3.57 1.7 Estimation of Error Terms Variance
table1.1 <- read.table(here('/data/CH01TA01.txt/'),header=FALSE) colnames(table1.1) <- c('lot.size','work.hours') mod <- lm(work.hours ~ 1 + lot.size,d=table1.1) table1.1$predicted <- predict(mod) table1.1$residuals <- resid(mod) table1.1$residuals.squared <- resid(mod)^2 sse = sum(table1.1$residuals.squared) sse
[1] 54825[1] 2384sqrt(mse)
[1] 48.821.8 Normal Error Regression Model
1.8.1 Least Square Estimation
table1.1 <- read.table(here('/data/CH01TA01.txt/'),header=FALSE) colnames(table1.1) <- c('lot.size','work.hours') beta0 <- seq(50,75,.1) beta1 <- seq(1,6,.01) ols <- expand.grid(beta0,beta1) colnames(ols) <- c('beta0','beta1') ols$ssr <- NA for(i in 1:nrow(ols)){ p = ols[i,1] + ols[i,2]*table1.1$lot.size ols[i,3] = sum((table1.1$work.hours - p)^2) } require(lattice) wireframe(ssr ~ beta0 * beta1, data = ols, shade=TRUE, screen = list(z = 40, x = -60, y=0), scales = list(arrows=FALSE), xlab = expression(beta[0]), ylab = expression(beta[1]), zlab = "SSR") ols[which.min(ols$ssr),]
beta0 beta1 ssr
64632 62.4 3.57 54825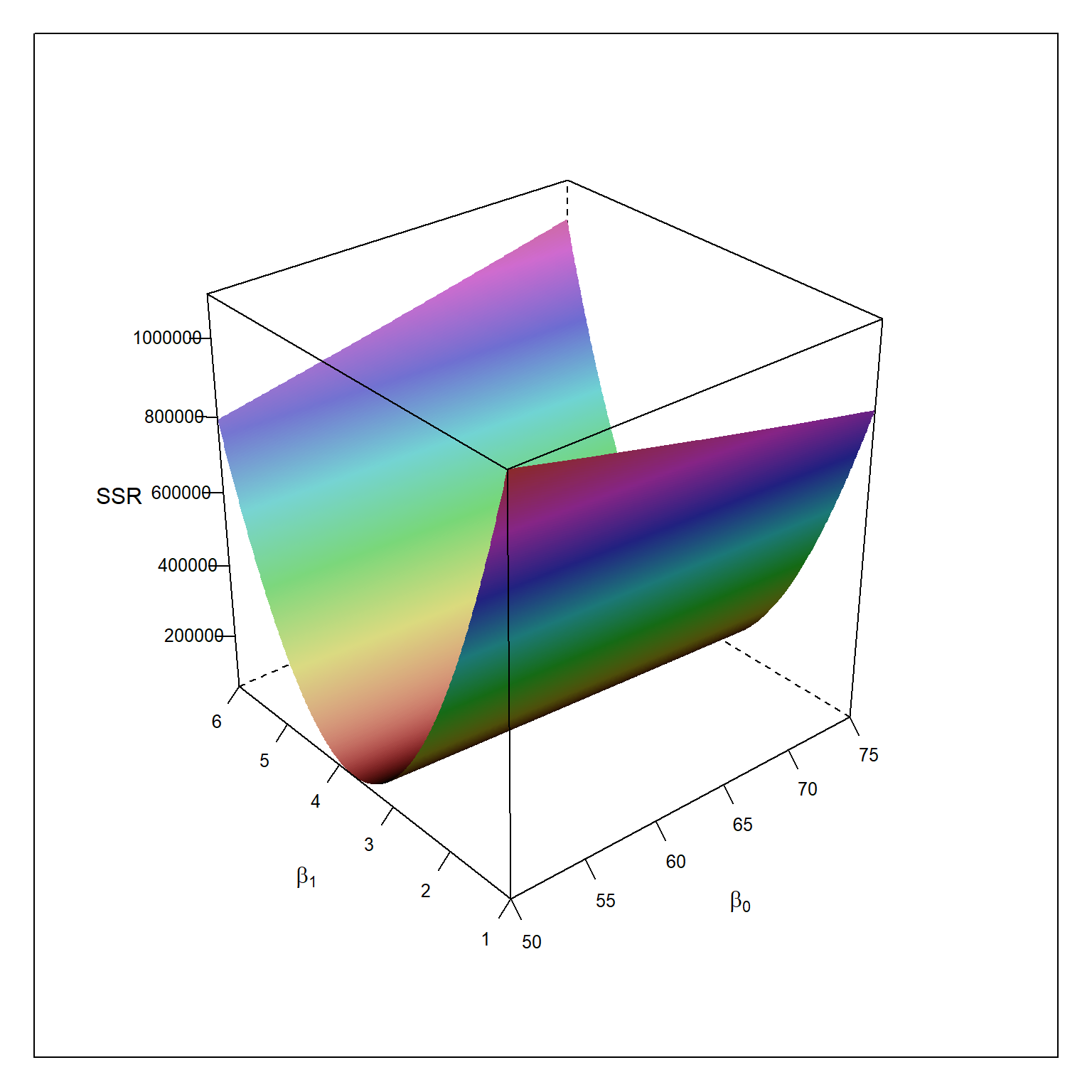
1.8.2 Maximum Likelihood Estimation
table1.1 <- read.table(here('/data/CH01TA01.txt/'),header=FALSE) colnames(table1.1) <- c('lot.size','work.hours') beta0 <- seq(50,75,.1) beta1 <- seq(1,6,.01) mle <- expand.grid(beta0,beta1) colnames(mle) <- c('beta0','beta1') mle$loglikelihood <- NA mse = 2383 # assumed to be known for(i in 1:nrow(mle)){ p = ols[i,1] + ols[i,2]*table1.1$lot.size mle[i,3] = sum(log(dnorm((table1.1$work.hours - p)/sqrt(mse)))) } wireframe(loglikelihood ~ beta0 * beta1, data = mle, shade=TRUE, screen = list(z = 40, x = -60, y=0), scales = list(arrows=FALSE), xlab = expression(beta[0]), ylab = expression(beta[1]), zlab = "Loglikelihood") mle[which.max(mle$loglikelihood),]
beta0 beta1 loglikelihood
64632 62.4 3.57 -34.48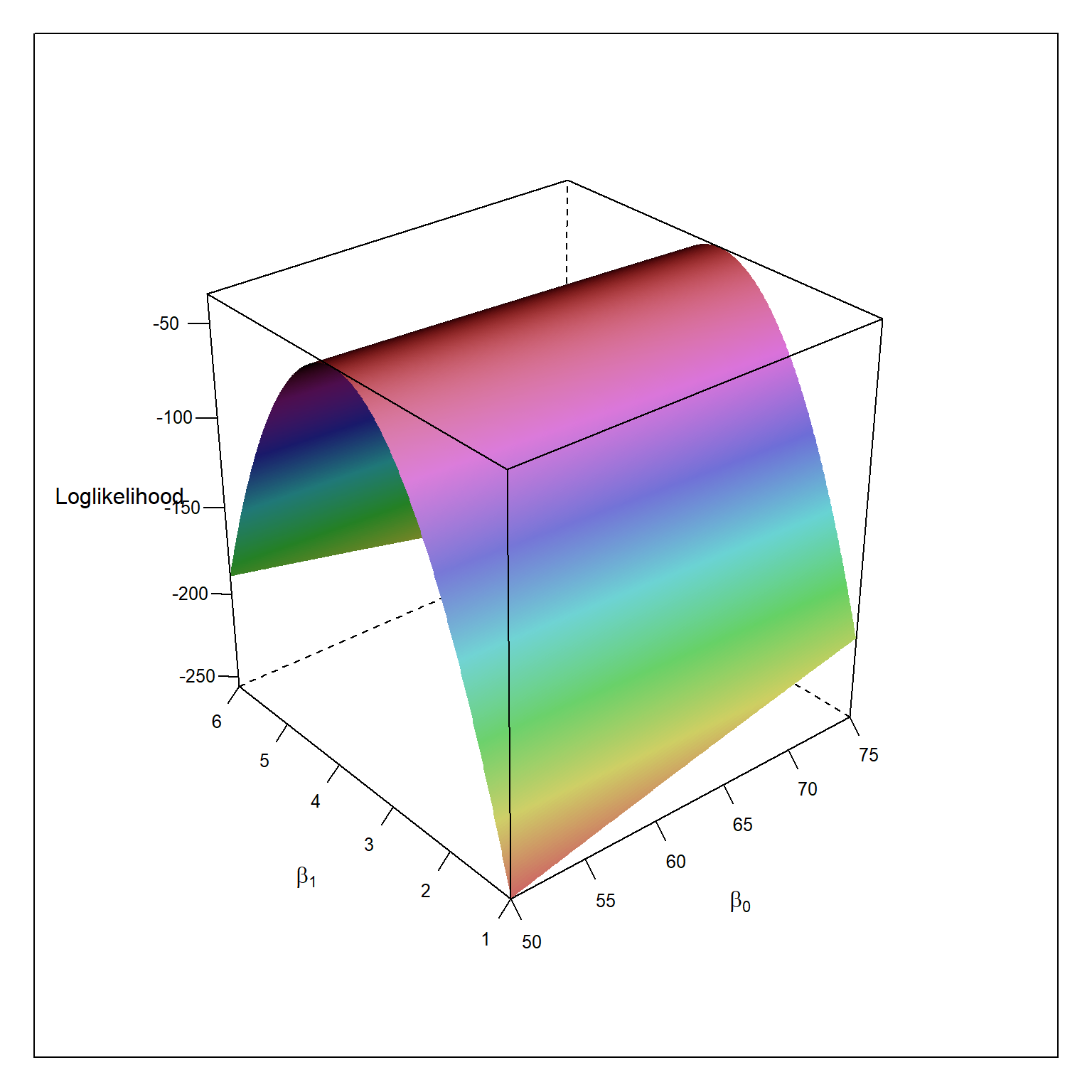
1.9 Problems
1.9.5 1.5
No. The expected value of Y should be the average, so there shouldn’t be an error part.
1.9.6 1.6
is the expected value of Y when X = 0, is the expected change in the value of Y for one unit increase in X
b0 = 200 b1 = 5 x = 0:70 mean = b0 + b1*x err.sd = 4 plot(x,mean,type="l", ylim=c(150,450), xlim=c(0,50), cex=1, pch=19, xlab="X", ylab="Y", xaxt='n') axis(side=1,at=c(10,20,40)) abline(b0,b1) dens = dnorm(seq(-3,3,.01),0,1) for(i in c(11,21,41)){ x. = x[i] - 5*dens y. = mean[i]+seq(-3,3,.01)*err.sd points(x.,y.,type="l",lty=2) abline(v=x[i],lty=2,col="gray") }
1.9.7 1.7
For , and . Assuming the errors are normally distributed, the probability that Y will be between 195 and 205 is about 68%.
1.9.8 1.8
No, the expected value for the new observations would be still 104 unless you re-fit the model with the addition of this new observation.
1.9.11 1.11
No, slope only shows the relationship between the production output before and after training. The intercept difference (20) reflects the training effect. So, training increased the production output by 20.
1.9.12 1.12
observational, no information that the senior citizens are randomly assigned to different levels of physical activity
not accurate, can’t make a causal statement
??
random assignment to different levels of physical activity
1.9.13 1.13
observational
not accurate, can’t make a causal statement
??
random assignment to different levels of physical activity
1.9.14 1.14
set.seed(1234) a <- data.frame(cbind(1:16,runif(16,0,1))) a <- a[order(a[,2]),] a$gr <- c(1,1,1,1,2,2,2,2,3,3,3,3,4,4,4,4) a
X1 X2 gr
7 7 0.009496 1
1 1 0.113703 1
8 8 0.232551 1
13 13 0.282734 1
15 15 0.292316 2
10 10 0.514251 2
12 12 0.544975 2
3 3 0.609275 2
2 2 0.622299 3
4 4 0.623379 3
6 6 0.640311 3
9 9 0.666084 3
11 11 0.693591 4
16 16 0.837296 4
5 5 0.860915 4
14 14 0.923433 4# Simulation code to demonstrate if the above procedure works as expected gr <- vector('list',100000) for(i in 1:100000){ a <- data.frame(cbind(1:16,runif(16,0,1))) a <- a[order(a[,2]),] a$gr <- c(1,1,1,1,2,2,2,2,3,3,3,3,4,4,4,4) gr[[i]] = a } assg <- c() k <- 4 for(i in 1:100000){ assg[i] <- gr[[i]][which(gr[[i]][,1]==k),3] } table(assg)
1.9.15 1.15
set.seed(1234) a <- data.frame(cbind(1:20,runif(20,0,1))) a <- a[order(a[,2]),] a$gr <- c(1,1,1,1,2,2,2,2,3,3,3,3,4,4,4,4,5,5,5,5) a
X1 X2 gr
7 7 0.009496 1
1 1 0.113703 1
19 19 0.186723 1
20 20 0.232226 1
8 8 0.232551 2
18 18 0.266821 2
13 13 0.282734 2
17 17 0.286223 2
15 15 0.292316 3
10 10 0.514251 3
12 12 0.544975 3
3 3 0.609275 3
2 2 0.622299 4
4 4 0.623379 4
6 6 0.640311 4
9 9 0.666084 4
11 11 0.693591 5
16 16 0.837296 5
5 5 0.860915 5
14 14 0.923433 51.9.16 1.16
No. Least squres method doesn’t care about the distribution of Y. It is still BLUE. No assumption is needed until one decides to make an inference
1.9.18 1.18
It is not necessary that the errors will sum to zero at the population. Even the average error in population is different than zero, the residuals from a regression at the sample level can be equal to zero. See below.
set.seed(1234) # Population b0 <- 200 b1 <- 5 sigma <- 5 x <- rnorm(10000,0,1) err <- rnorm(10000,50,5) y <- b0+b1+err # A random sample with size 200 s <- sample(1:10000,200) s.x <- x[s] s.y <- y[s] fit <- lm(s.y ~ s.x) summary(fit)
Call:
lm(formula = s.y ~ s.x)
Residuals:
Min 1Q Median 3Q Max
-17.404 -2.695 0.236 3.044 13.149
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 254.808 0.346 735.87 <0.0000000000000002 ***
s.x 0.299 0.350 0.85 0.39
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 4.89 on 198 degrees of freedom
Multiple R-squared: 0.00366, Adjusted R-squared: -0.00137
F-statistic: 0.727 on 1 and 198 DF, p-value: 0.395[1] -0.000000000000000073381.9.19 1.19
require(here) gpa <- read.table(here('data/CH01PR19.txt'), header=FALSE, col.names = c('gpa','act')) str(gpa)
'data.frame': 120 obs. of 2 variables:
$ gpa: num 3.9 3.88 3.78 2.54 3.03 ...
$ act: int 21 14 28 22 21 31 32 27 29 26 ...head(gpa)
gpa act
1 3.897 21
2 3.885 14
3 3.778 28
4 2.540 22
5 3.028 21
6 3.865 31(Intercept) act
2.11405 0.03883 summary(fit)
Call:
lm(formula = gpa ~ act, data = gpa)
Residuals:
Min 1Q Median 3Q Max
-2.7400 -0.3383 0.0406 0.4406 1.2274
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 2.1140 0.3209 6.59 0.0000000013 ***
act 0.0388 0.0128 3.04 0.0029 **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.623 on 118 degrees of freedom
Multiple R-squared: 0.0726, Adjusted R-squared: 0.0648
F-statistic: 9.24 on 1 and 118 DF, p-value: 0.00292require(ggplot2) ggplot(data = gpa, aes(x=act,y=gpa))+ geom_point()+ geom_function(fun = function(x) coef(fit)[1] + coef(fit)[2]*x)+ theme_bw()
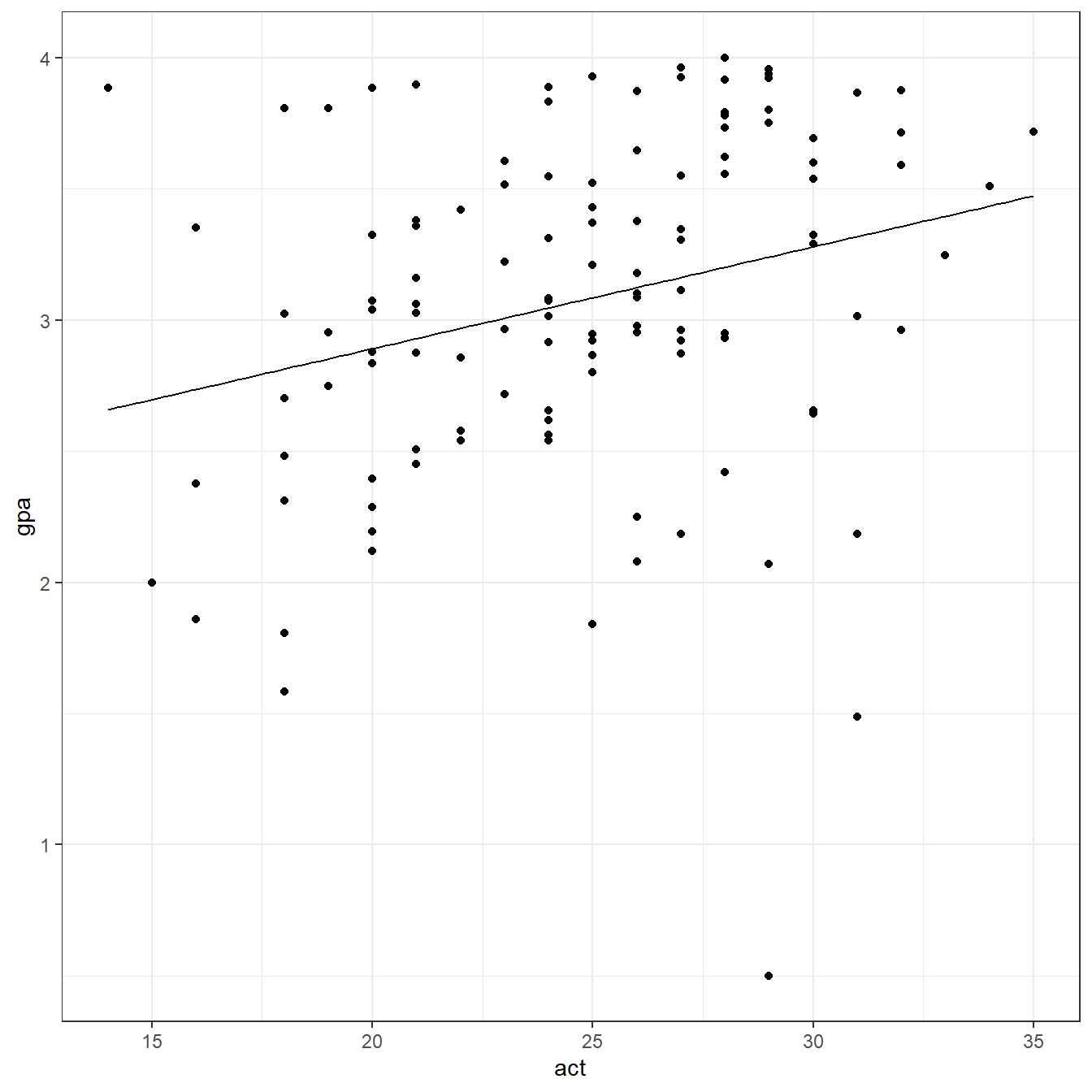
predict(fit)
1 2 3 4 5 6 7 8 9 10 11 12 13
2.929 2.658 3.201 2.968 2.929 3.318 3.357 3.162 3.240 3.124 3.046 3.279 3.046
14 15 16 17 18 19 20 21 22 23 24 25 26
3.046 3.395 3.162 3.085 3.318 3.085 2.891 3.046 2.929 3.201 3.162 3.201 3.124
27 28 29 30 31 32 33 34 35 36 37 38 39
3.201 2.968 3.124 2.929 3.085 2.735 3.201 3.124 2.968 3.046 2.929 3.279 3.162
40 41 42 43 44 45 46 47 48 49 50 51 52
3.124 3.124 3.279 3.046 3.124 3.240 3.046 3.318 2.696 2.852 2.813 3.162 2.735
53 54 55 56 57 58 59 60 61 62 63 64 65
3.162 3.124 3.046 3.279 2.929 2.891 3.279 3.240 3.085 3.007 3.085 3.007 3.279
66 67 68 69 70 71 72 73 74 75 76 77 78
2.929 3.046 3.357 2.813 3.007 2.891 3.007 2.813 2.813 3.240 2.891 3.007 3.124
79 80 81 82 83 84 85 86 87 88 89 90 91
3.201 3.434 2.891 2.891 3.124 3.357 3.085 3.162 3.162 3.240 2.852 2.929 3.046
92 93 94 95 96 97 98 99 100 101 102 103 104
3.162 3.085 2.813 3.240 3.046 3.162 2.929 2.852 2.813 3.085 2.813 2.891 3.357
105 106 107 108 109 110 111 112 113 114 115 116 117
3.046 3.473 3.085 3.201 3.201 3.085 2.968 3.279 2.891 2.891 3.318 2.891 3.240
118 119 120
3.201 2.735 3.201 resid(fit)
1 2 3 4 5 6 7 8
0.96758 1.22737 0.57679 -0.42825 0.09858 0.54731 -0.39452 0.79862
9 10 11 12 13 14 15 16
-2.74004 0.05445 0.26410 0.25914 0.03710 -0.03290 -0.15034 -0.19938
17 18 19 20 21 22 23 24
0.43727 -0.30469 -0.13773 -0.77259 -0.48290 0.42758 0.52979 0.76262
25 26 27 28 29 30 31 32
0.35479 -0.02255 -0.78121 -0.38925 0.74745 0.13058 0.84227 -0.36028
33 34 35 36 37 38 39 40
-0.27221 0.25145 -0.11125 0.02610 0.45158 0.01114 0.38662 0.52245
41 42 43 44 45 46 47 48
-0.14555 -0.62486 -0.50590 -0.87355 -1.17104 -0.42890 -1.13469 -0.69646
49 50 51 52 53 54 55 56
0.10024 0.99306 -0.29138 0.61672 0.14262 -0.17155 0.50110 0.41214
57 58 59 60 61 62 63 64
0.23058 -0.69659 0.04414 0.69596 -0.16273 -0.29107 0.28527 0.59893
65 66 67 68 69 70 71 72
-0.63686 -0.47742 -0.39090 0.35748 -1.00694 0.50893 0.14841 -0.04107
73 74 75 76 77 78 79 80
-0.33094 -0.11294 0.67996 -0.05659 0.21493 -0.03955 0.79879 0.07683
81 82 83 84 85 86 87 88
0.43241 0.18141 -1.04455 0.51848 0.12327 -0.24238 0.18262 0.71596
89 90 91 92 93 94 95 96
0.95624 -0.42342 0.84010 -0.97938 0.34427 0.21106 0.50996 0.78710
97 98 99 100 101 102 103 104
-0.04938 -0.05442 -0.10476 -0.50194 -1.24373 -1.22994 -0.01159 0.23448
105 106 107 108 109 110 111 112
-0.13190 0.24300 -0.28473 0.41979 0.59079 -0.21773 0.45075 0.32114
113 114 115 116 117 118 119 120
-0.49659 -0.60459 -1.83169 0.99441 0.55996 0.71279 -0.87528 -0.25321 [1] -0.00000000000000002453[1] 0.385[1] 0.6205predict(fit, newdata = data.frame(act = 30))
1
3.279 1.9.20 1.20
copier <- read.table(here('data/CH01PR20.txt'), header=FALSE, col.names = c('y','x')) fit <- lm(y ~ x, data=copier) coef(fit)
(Intercept) x
-0.5802 15.0352 summary(fit)
Call:
lm(formula = y ~ x, data = copier)
Residuals:
Min 1Q Median 3Q Max
-22.772 -3.737 0.333 6.333 15.404
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -0.580 2.804 -0.21 0.84
x 15.035 0.483 31.12 <0.0000000000000002 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 8.91 on 43 degrees of freedom
Multiple R-squared: 0.957, Adjusted R-squared: 0.957
F-statistic: 969 on 1 and 43 DF, p-value: <0.0000000000000002ggplot(data = copier, aes(x=x,y=y))+ geom_point()+ geom_function(fun = function(x) coef(fit)[1] + coef(fit)[2]*x)+ theme_bw()
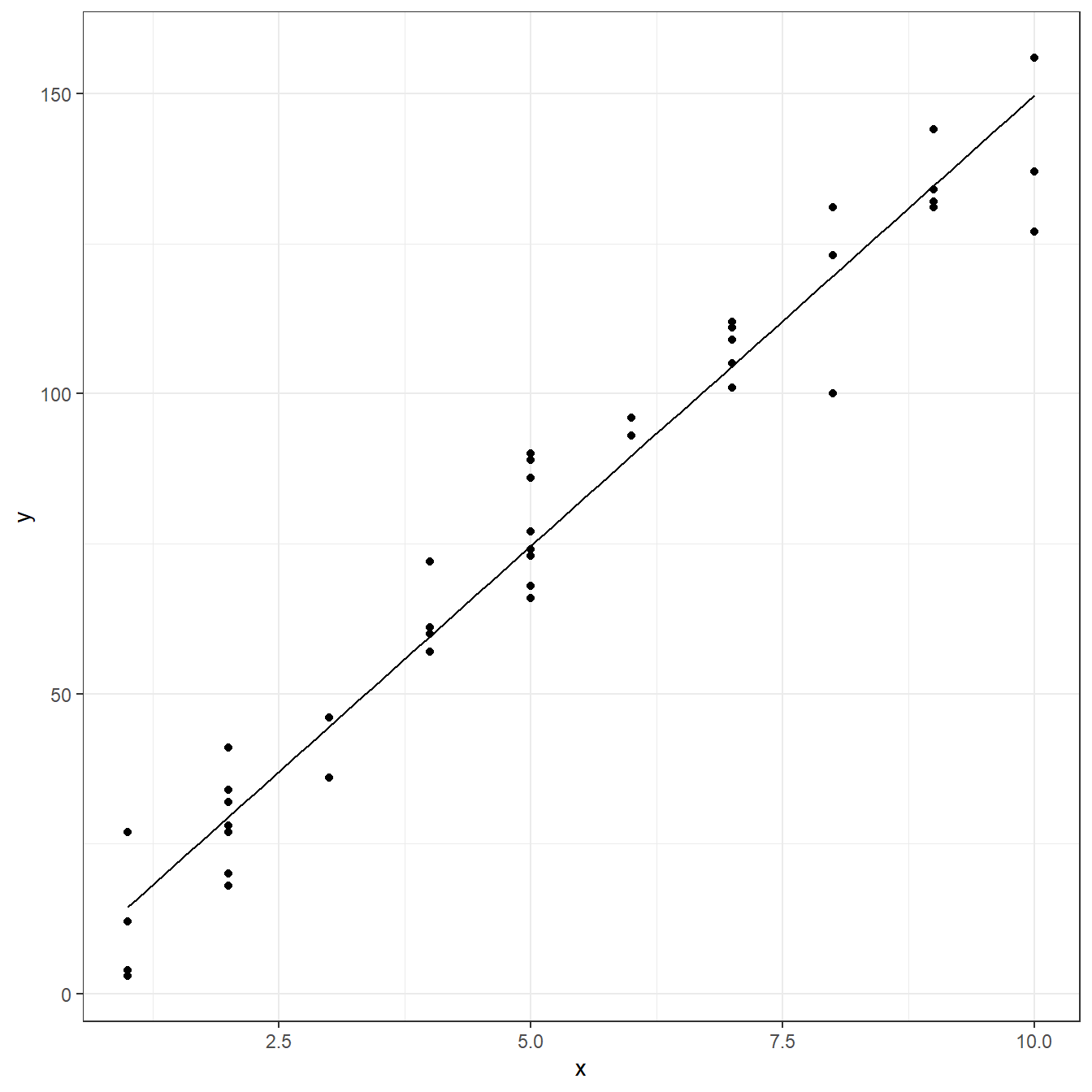
predict(fit)
1 2 3 4 5 6 7 8 9 10 11
29.49 59.56 44.53 29.49 14.46 149.77 74.60 74.60 14.46 29.49 134.74
12 13 14 15 16 17 18 19 20 21 22
149.77 89.63 44.53 59.56 119.70 104.67 119.70 149.77 59.56 74.60 104.67
23 24 25 26 27 28 29 30 31 32 33
104.67 74.60 134.74 104.67 29.49 74.60 104.67 89.63 119.70 74.60 29.49
34 35 36 37 38 39 40 41 42 43 44
29.49 14.46 59.56 74.60 134.74 104.67 14.46 134.74 29.49 29.49 59.56
45
74.60 resid(fit)
1 2 3 4 5 6 7 8
-9.4903 0.4392 1.4744 11.5097 -2.4551 -12.7723 -6.5961 14.4039
9 10 11 12 13 14 15 16
-10.4551 2.5097 9.2629 6.2277 3.3687 -8.5256 12.4392 -19.7018
17 18 19 20 21 22 23 24
0.3334 11.2982 -22.7723 -2.5608 -8.5961 -3.6666 4.3334 -0.5961
25 26 27 28 29 30 31 32
-0.7371 7.3334 -11.4903 -1.5961 6.3334 6.3687 3.2982 15.4039
33 34 35 36 37 38 39 40
-9.4903 -1.4903 -11.4551 -2.5608 11.4039 -2.7371 7.3334 12.5449
41 42 43 44 45
-3.7371 4.5097 -2.4903 1.4392 2.4039 [1] -0.0000000000000002612[1] 77.64[1] 8.812predict(fit, newdata = data.frame(x = 5))
1
74.6 1.9.21 1.21
freight <- read.table(here('data/CH01PR21.txt'), header=FALSE, col.names = c('y','x')) fit <- lm(y ~ x, data=freight) coef(fit)
(Intercept) x
10.2 4.0 summary(fit)
Call:
lm(formula = y ~ x, data = freight)
Residuals:
Min 1Q Median 3Q Max
-2.2 -1.2 0.3 0.8 1.8
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 10.200 0.663 15.38 0.00000032 ***
x 4.000 0.469 8.53 0.00002749 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 1.48 on 8 degrees of freedom
Multiple R-squared: 0.901, Adjusted R-squared: 0.889
F-statistic: 72.7 on 1 and 8 DF, p-value: 0.0000275ggplot(data = freight, aes(x=x,y=y))+ geom_point()+ geom_function(fun = function(x) coef(fit)[1] + coef(fit)[2]*x)+ theme_bw()
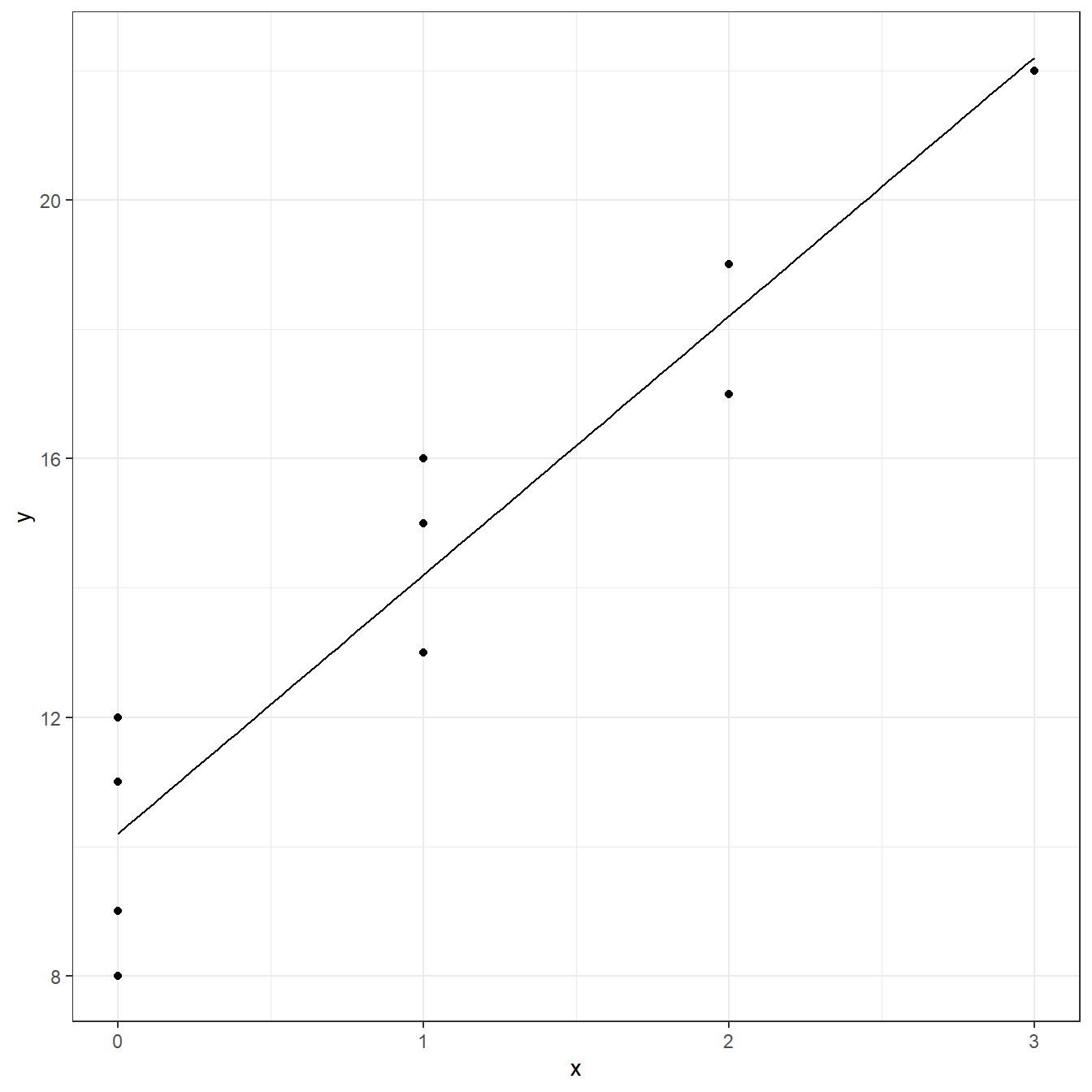
predict(fit)
1 2 3 4 5 6 7 8 9 10
14.2 10.2 18.2 10.2 22.2 14.2 10.2 14.2 18.2 10.2 resid(fit)
1 2 3 4 5 6 7 8 9 10
1.8 -1.2 -1.2 1.8 -0.2 -1.2 -2.2 0.8 0.8 0.8 [1] -0.00000000000000004995[1] 1.956[1] 1.398predict(fit, newdata = data.frame(x = 1))
1
14.2 1.9.22 1.22
hardness <- read.table(here('data/CH01PR22.txt'), header=FALSE, col.names = c('y','x')) fit <- lm(y ~ x, data=hardness) coef(fit)
(Intercept) x
168.600 2.034 summary(fit)
Call:
lm(formula = y ~ x, data = hardness)
Residuals:
Min 1Q Median 3Q Max
-5.150 -2.219 0.162 2.688 5.575
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 168.6000 2.6570 63.5 < 0.0000000000000002 ***
x 2.0344 0.0904 22.5 0.0000000000022 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 3.23 on 14 degrees of freedom
Multiple R-squared: 0.973, Adjusted R-squared: 0.971
F-statistic: 507 on 1 and 14 DF, p-value: 0.00000000000216ggplot(data = hardness, aes(x=x,y=y))+ geom_point()+ geom_function(fun = function(x) coef(fit)[1] + coef(fit)[2]*x)+ theme_bw()
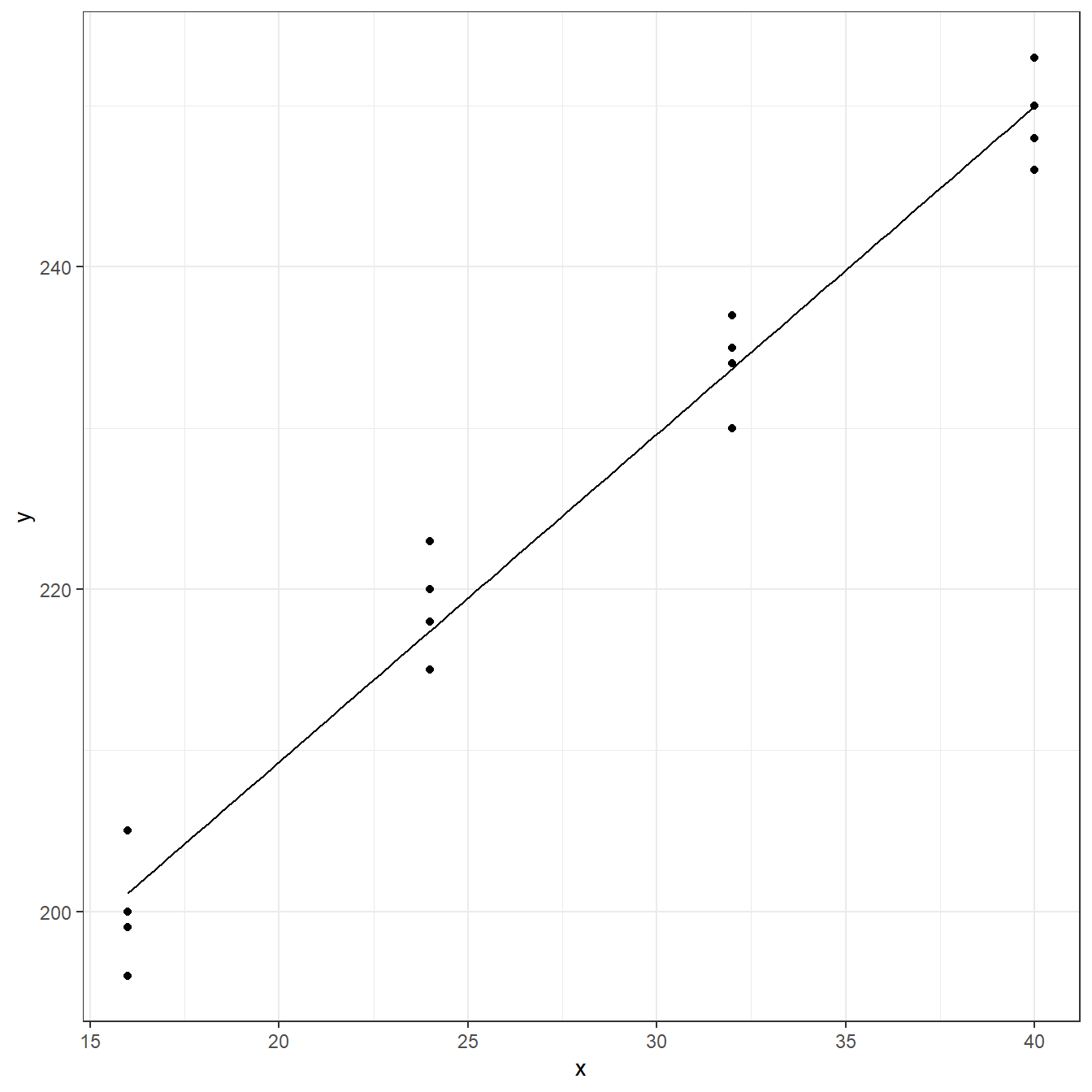
predict(fit)
1 2 3 4 5 6 7 8 9 10 11 12 13
201.1 201.1 201.1 201.1 217.4 217.4 217.4 217.4 233.7 233.7 233.7 233.7 250.0
14 15 16
250.0 250.0 250.0 resid(fit)
1 2 3 4 5 6 7 8 9 10 11
-2.150 3.850 -5.150 -1.150 0.575 2.575 -2.425 5.575 3.300 0.300 1.300
12 13 14 15 16
-3.700 0.025 -1.975 3.025 -3.975 [1] -0.0000000000000001249[1] 9.762[1] 3.124predict(fit, newdata = data.frame(x = 40))
1
250 1.9.27 1.27
muscle <- read.table(here('data/CH01PR27.txt'), header=FALSE, col.names = c('y','x')) fit <- lm(y ~ x, data=muscle) coef(fit)
(Intercept) x
156.35 -1.19 summary(fit)
Call:
lm(formula = y ~ x, data = muscle)
Residuals:
Min 1Q Median 3Q Max
-16.137 -6.197 -0.597 6.761 23.473
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 156.3466 5.5123 28.4 <0.0000000000000002 ***
x -1.1900 0.0902 -13.2 <0.0000000000000002 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 8.17 on 58 degrees of freedom
Multiple R-squared: 0.75, Adjusted R-squared: 0.746
F-statistic: 174 on 1 and 58 DF, p-value: <0.0000000000000002ggplot(data = muscle, aes(x=x,y=y))+ geom_point()+ geom_function(fun = function(x) coef(fit)[1] + coef(fit)[2]*x)+ theme_bw()
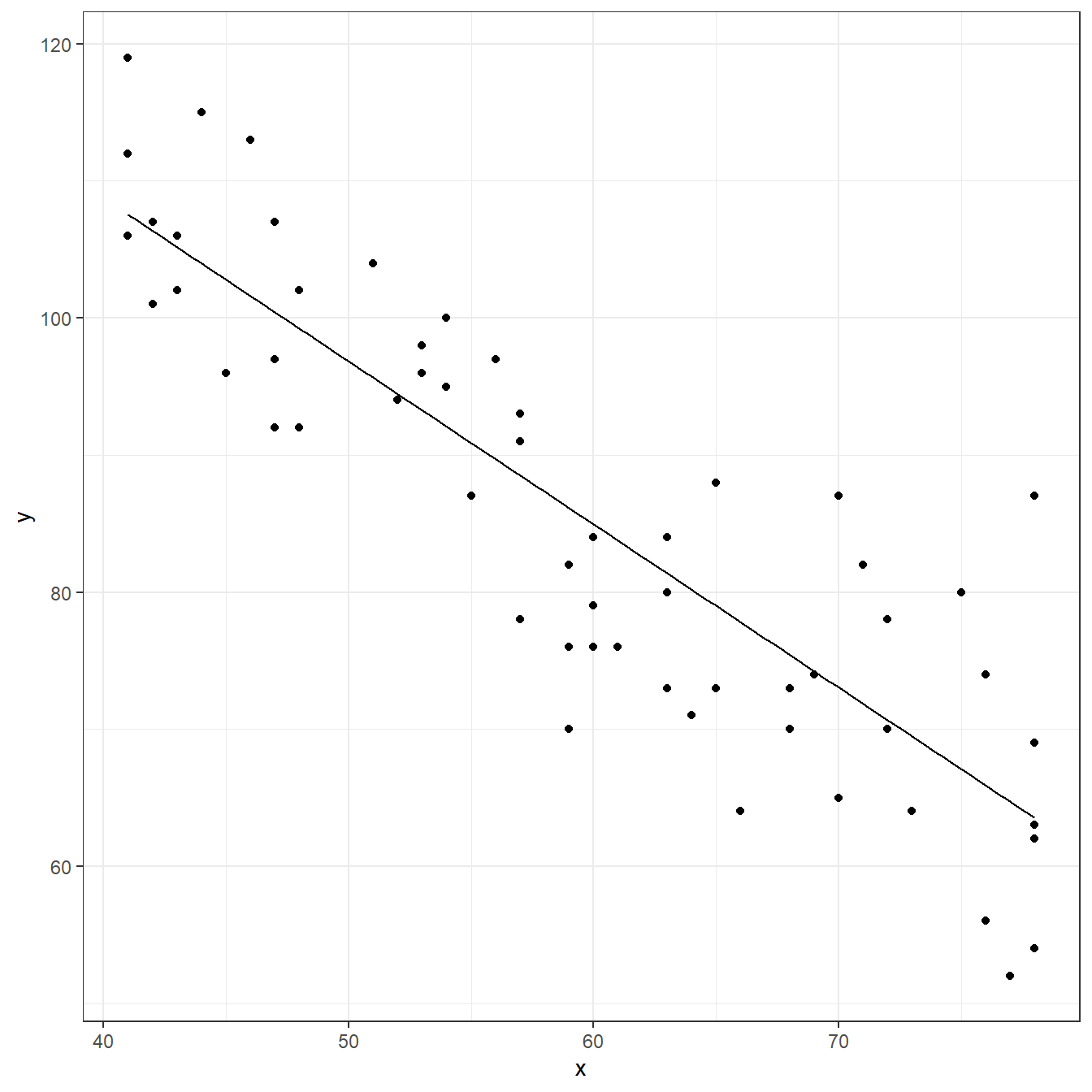
predict(fit)
1 2 3 4 5 6 7 8 9 10 11
105.18 107.56 100.42 101.61 102.80 107.56 100.42 107.56 99.23 99.23 106.37
12 13 14 15 16 17 18 19 20 21 22
100.42 105.18 103.99 106.37 90.90 88.52 89.71 86.14 88.52 92.09 93.28
23 24 25 26 27 28 29 30 31 32 33
94.47 93.28 92.09 84.95 86.14 95.66 86.14 88.52 75.43 81.38 84.95
34 35 36 37 38 39 40 41 42 43 44
81.38 81.38 80.19 77.81 79.00 84.95 79.00 79.00 74.24 83.76 73.05
45 46 47 48 49 50 51 52 53 54 55
75.43 63.53 63.53 63.53 70.67 73.05 69.48 65.91 63.53 63.53 71.86
56 57 58 59 60
67.10 64.72 65.91 70.67 65.91 resid(fit)
1 2 3 4 5 6 7 8
0.8232 -1.5567 -3.4168 11.3932 -6.7968 11.4433 -8.4168 4.4433
9 10 11 12 13 14 15 16
-7.2268 2.7732 0.6332 6.5832 -3.1768 11.0132 -5.3668 -3.8968
17 18 19 20 21 22 23 24
2.4832 7.2932 -4.1368 -10.5168 2.9132 4.7232 -0.4668 2.7232
25 26 27 28 29 30 31 32
7.9132 -0.9468 -16.1368 8.3432 -10.1368 4.4832 -2.4269 -8.3768
33 34 35 36 37 38 39 40
-8.9468 -1.3768 2.6232 -9.1869 -13.8069 9.0031 -5.9468 9.0031
41 42 43 44 45 46 47 48
-5.9969 -0.2369 -7.7568 13.9531 -5.4269 5.4731 -9.5269 -1.5269
49 50 51 52 53 54 55 56
7.3331 -8.0469 -5.4769 8.0931 23.4731 -0.5269 10.1431 12.9031
57 58 59 60
-12.7169 -9.9069 -0.6669 8.0931 [1] 0.00000000000000001123[1] 65.67[1] 8.104predict(fit, newdata = data.frame(x = 40))
1
108.7 1.9.28 1.28
crime <- read.table(here('data/CH01PR28.txt'), header=FALSE, col.names = c('crime','edu')) fit <- lm(crime ~ edu, data=crime) coef(fit)
(Intercept) edu
20517.6 -170.6 summary(fit)
Call:
lm(formula = crime ~ edu, data = crime)
Residuals:
Min 1Q Median 3Q Max
-5278 -1758 -210 1575 6803
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 20517.6 3277.6 6.26 0.000000017 ***
edu -170.6 41.6 -4.10 0.000095714 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 2360 on 82 degrees of freedom
Multiple R-squared: 0.17, Adjusted R-squared: 0.16
F-statistic: 16.8 on 1 and 82 DF, p-value: 0.0000957ggplot(data = crime, aes(x=edu,y=crime))+ geom_point()+ geom_function(fun = function(x) coef(fit)[1] + coef(fit)[2]*x)+ theme_bw()
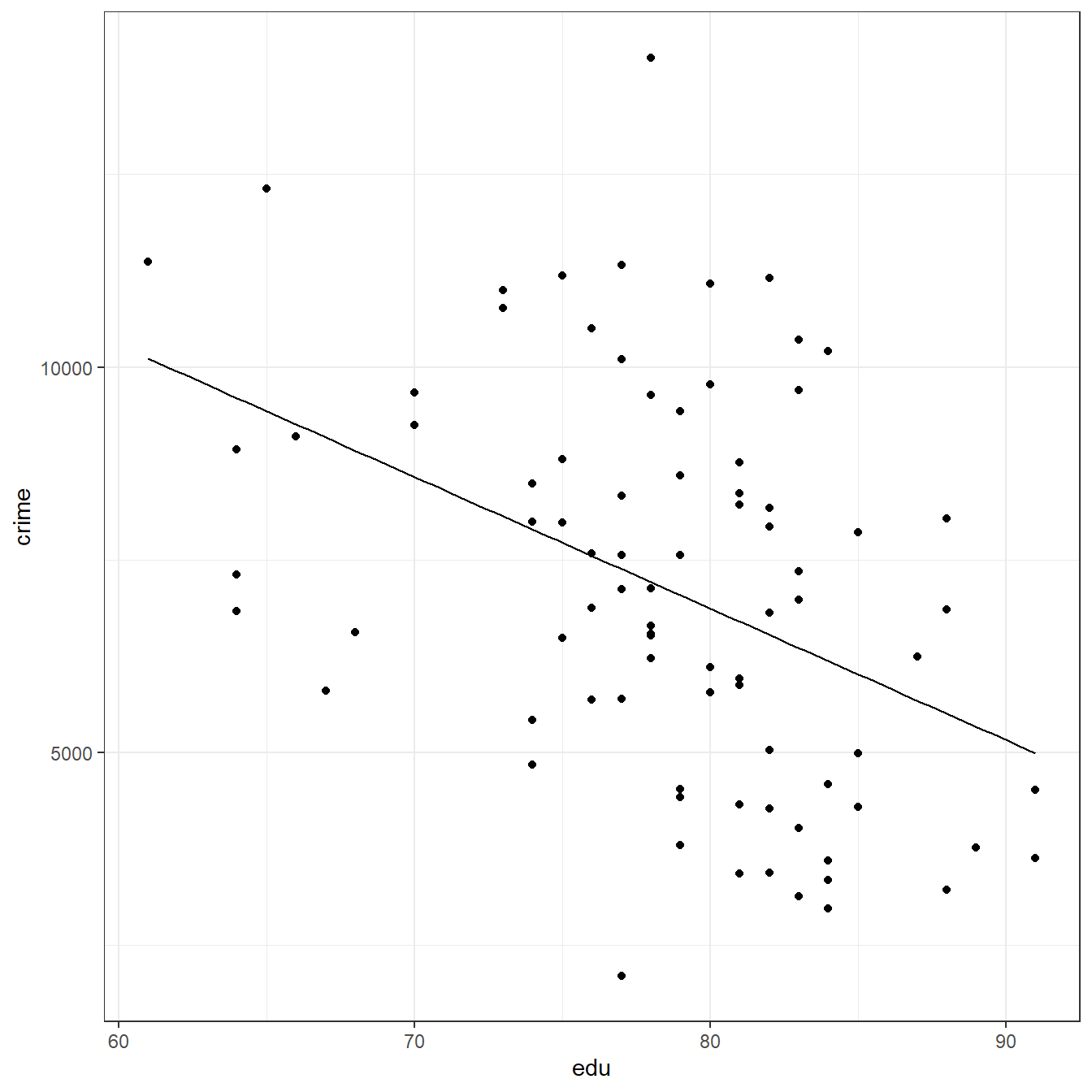
predict(fit)
1 2 3 4 5 6 7 8 9 10 11 12 13
7895 6530 6701 6701 5678 9260 8918 6701 7895 6530 7724 6530 7213
14 15 16 17 18 19 20 21 22 23 24 25 26
6189 6530 7042 7213 8066 7383 9430 7383 7554 7042 7042 7213 6189
27 28 29 30 31 32 33 34 35 36 37 38 39
7213 6701 5336 6019 7383 7895 6872 6189 5507 7724 7383 7213 10113
40 41 42 43 44 45 46 47 48 49 50 51 52
4995 6360 7383 6019 8577 5507 6872 6530 6530 6530 8577 9601 7042
53 54 55 56 57 58 59 60 61 62 63 64 65
6360 7383 7554 6872 6189 6530 6701 7895 6701 7554 7213 7213 7042
66 67 68 69 70 71 72 73 74 75 76 77 78
6360 7042 6360 6701 6189 9601 9089 7724 8066 7383 9601 7724 6872
79 80 81 82 83 84
6360 6019 4995 5507 6360 7554 resid(fit)
1 2 3 4 5 6 7 8
591.96 1648.57 1660.99 1518.99 568.44 -159.64 -2357.49 -828.01
9 10 11 12 13 14 15 16
97.96 1401.57 -1233.46 285.57 2426.26 -1594.28 -1493.43 -2615.16
17 18 19 20 21 22 23 24
-986.74 2702.39 951.69 2880.79 2720.69 2949.11 519.84 1550.84
25 26 27 28 29 30 31 32
-79.74 4015.72 6803.26 -742.01 -1572.41 -1721.71 178.69 -3051.04
33 34 35 36 37 38 39 40
-1094.58 -2590.28 -2287.98 3462.54 -5278.31 -562.74 1258.49 -478.26
41 42 43 44 45 46 47 48
988.14 -1687.31 -1023.71 670.66 1353.02 2904.42 -2250.43 4623.57
49 50 51 52 53 54 55 56
-3088.43 1096.66 -2291.79 -2512.16 -2342.86 -261.31 -1864.89 -762.58
57 58 59 60 61 62 63 64
-2846.28 -1501.43 -2371.01 -2470.04 2067.99 -673.89 -674.74 -691.74
65 66 67 68 69 70 71 72
2380.84 3337.14 -3237.16 -3225.86 -3268.01 -3210.28 -2764.79 -3285.06
73 74 75 76 77 78 79 80
261.54 2928.39 3938.69 -663.79 1082.54 4215.42 3995.14 1839.29
81 82 83 84
-1363.26 2533.02 621.14 28.11 [1] 0.00000000000001277[1] 5485219[1] 2342predict(fit, newdata = data.frame(edu = 80))
1
6872